Atopic dermatitis linked to the same parts of the genome in dogs and humans
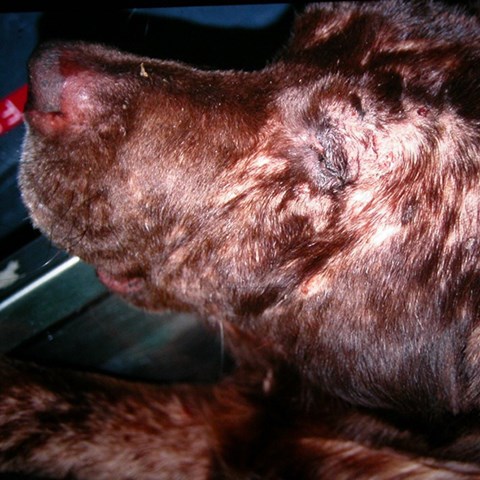
Using new gene mapping methods, researchers have found connections between atopic dermatitis (eczema) in dogs and several regions of the genome. Some of the genes identified coincide with genes linked to similar problems in humans. The filaggrin gene region, for example, which is regarded as the most powerful risk factor for atopic eczema in humans, has now also been linked to this disease in Labrador retrievers.
The findings are reported in a new study produced by the dog genetics group at Uppsala University and the Swedish University of Agricultural Sciences, which has been conducting research in this field for more than ten years in cooperation with colleagues in Switzerland, the United Kingdom and the United States.
Genome sequencing became possible in the early-2000s. Since then, canine genome sequencing has proved invaluable for researchers seeking to understand the human genome.
Dogs have lived alongside humans for tens of thousands of years and suffer from similar diseases, including immunological diseases such as atopic dermatitis (allergic eczema). Studying dog disease genetics, using ordinary blood samples, can also be a way to obtain clues to the causes of the corresponding human diseases.
Dogs and humans affected by atopic eczema have much in common as regards medical symptoms and the early onset of the disease, as well as in histopathological terms with similar immune cell infiltration in the skin. The disease has a complex genetic background in both species and is also influenced by environmental factors.
“It’s vital that atopic eczema is correctly diagnosed by careful elimination of other potential non-allergic causes of the patient’s symptoms, followed by a positive allergy test,” says Kerstin Bergvall, veterinarian in charge and specialist in dermatology at the Swedish University of Agricultural Sciences, who has been involved in the research from the start.
With gene mapping technology steadily advancing, new methods have recently emerged for mapping complex diseases. In the study now published in the journal Communications Biology, the researchers used one methodology to capture multiple associated genetic risk variants and another to discover disease variants ‘hidden’ in the genome because of artificially (i.e. humanly) selected characteristics.
“The new methods make it possible to find new risk factors that have become common in the specific breed, perhaps because of the selection for other characteristics,” says Katarina Tengvall, researcher at Uppsala University and first author of the study. “The candidate genes identified here are important for both the nature of the skin barrier and the immune defence, as expected in atopic eczema.”
The study shows several overlaps, or correspondences, with genes associated with human atopic dermatitis. One particularly striking finding now made by the researchers in this study is that the genetic region containing the filaggrin gene, which is regarded as the most powerful genetic risk factor for atopic eczema in humans, is a risk factor in dogs as well.
“This highlights the value of canine studies of genetic diseases that also affect humans. A better understanding of the disease mechanisms may ultimately lead to better therapies for dog and human alike,” says Kerstin Lindblad-Toh, Professor of Comparative Genomics and senior author of the study.
This text is based on a press release from Uppsala University.
The article
Tengvall et al. (2022); Bayesian model and selection signature analyses reveal risk factors for canine atopic dermatitis, Communications Biology, DOI : 10.1038/s42003-022-04279-8. https://doi.org/10.1038/s42003-022-04279-8
Contact persons
Katarina Tengvall, researcher at the Department of Medical Biochemistry and Microbiology, Uppsala University, katarina.tengvall@imbim.uu.se
Kerstin Lindblad-Toh, Professor of Comparative Genomics at the Department of Medical Biochemistry and Microbiology, Uppsala University, kersli@broadinstitute.org
Kerstin Bergvall: veterinarian at the Department of Clinical Sciences, Swedish University of Agricultural Sciences, kerstin.bergvall@slu.se
Göran Andersson: professor of molecular genetics at the Department of Animal Breeding and Genetics, Swedish University of Agricultural Sciences, goran.andersson@slu.se
Major funding for the research was provided by the European Research Council (ERC) and the Swedish Research Council (VR).